Biosensors for biomass-derived building blocks: Modern society is hugely dependent on finite oil reserves for the supply of fuels and chemicals. Moving our dependence away from these unsustainable oil-based feedstocks to renewable ones is therefore a critical factor towards the development of a low carbon bioeconomy. Lignin derived from biomass feedstocks offers great potential as a renewable source of aromatic compounds if methods for its effective valorization can be developed. Genetically encoded biosensors can provide enabling tools by transducing the target metabolite concentration into detectable signals to provide high-throughput phenotypic read-outs and allow dynamic pathway regulation (Biotechnology for Biofuels 2019).
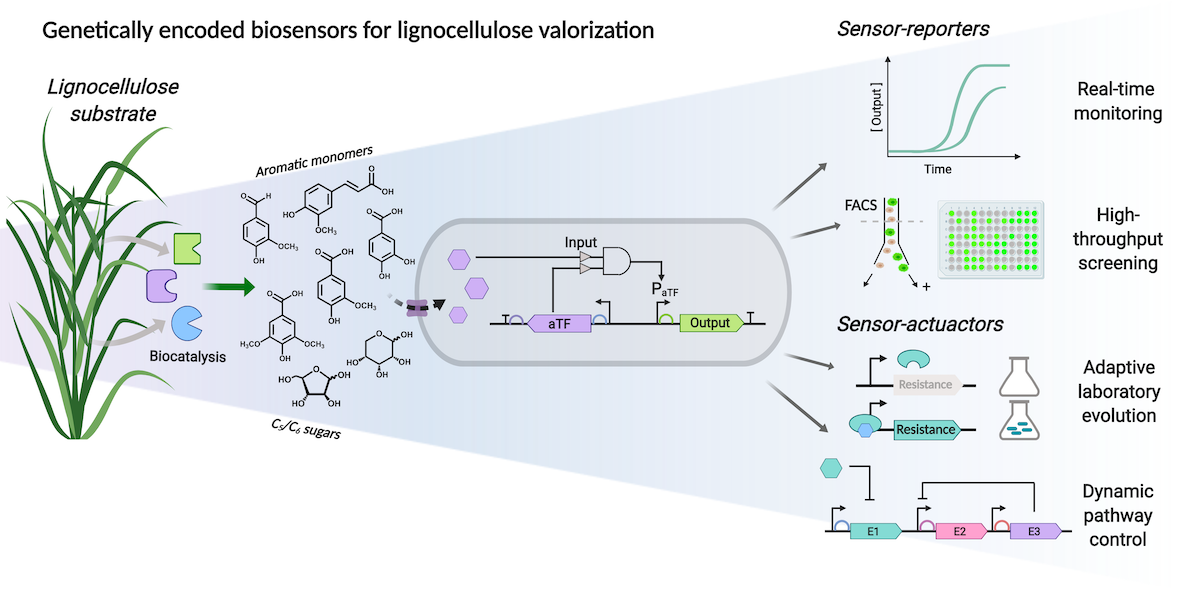
Biosensors for lignin-derived building blocks: We previously developed a biosensor that permits the detection of aromatic building blocks (e.g ferulic acid) derived from lignin biomass. This system operates via a repression depression mechanism utilising a marR aTF that results in the expression of a reporter gene (eGFP) in the presence of substrate. We are currently working with researchers in Brazil, and at the University of Warwick, to exploit this technology for the degradation and valorisation of sugarcane bagasse (Chem Commun 2016).
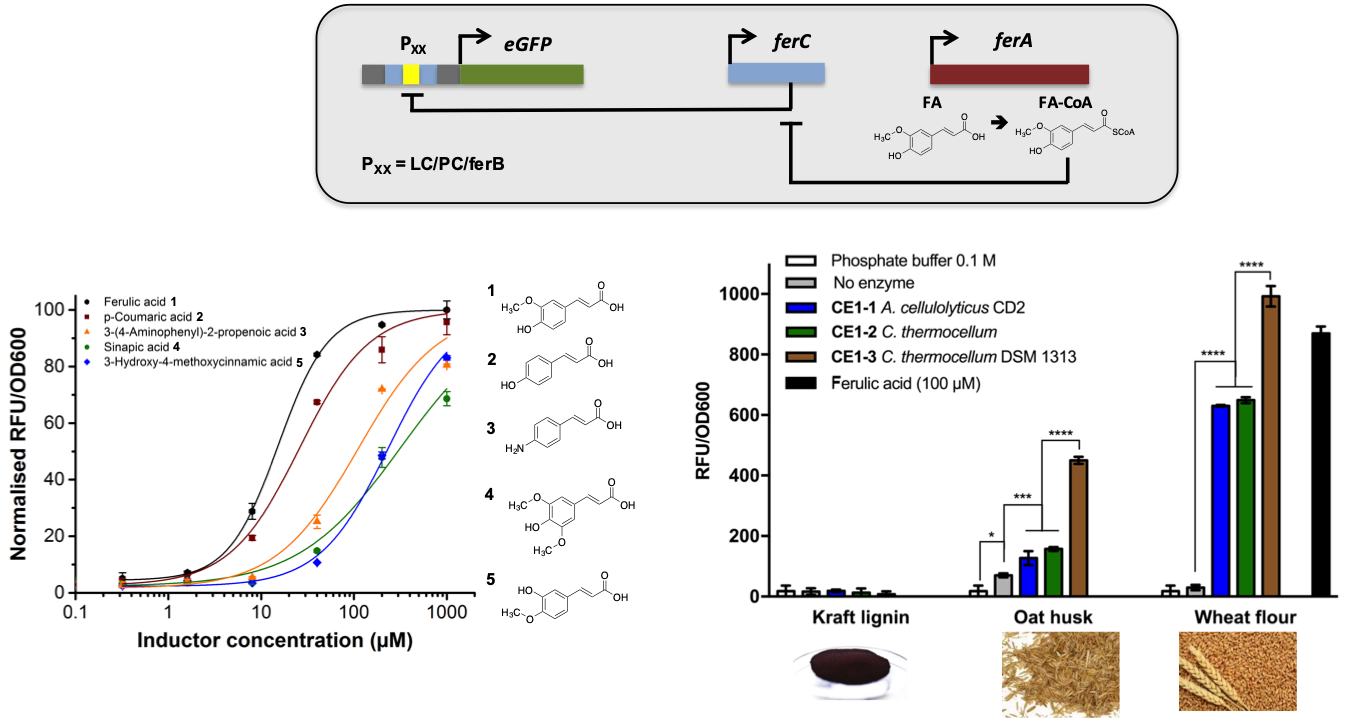
Design of Experiments for biosensor circuit optimisation: The gene expression level of biosensor regulatory components required for optimal performance is nonintuitive, and classical iterative approaches do not efficiently explore multidimensional experimental space. To overcome these challenges, we proposed and implemented a design of experiments (DoE) methodology to efficiently map gene expression levels and provide biosensors with enhanced performance. This methodology was applied to two biosensors that respond to catabolic breakdown products of lignin biomass, protocatechuic acid and ferulic acid. Utilizing DoE we systematically modified biosensor dose–response behavior by increasing the maximum signal output (up to 30-fold increase), improving dynamic range (>500-fold), expanding the sensing range (∼4-orders of magnitude), increasing sensitivity (by >1500-fold), and modulated the slope of the curve to afford biosensors designs with both digital and analogue dose–response behavior. This DoE method shows promise for the optimization of regulatory systems and metabolic pathways constructed from novel, poorly characterized parts.
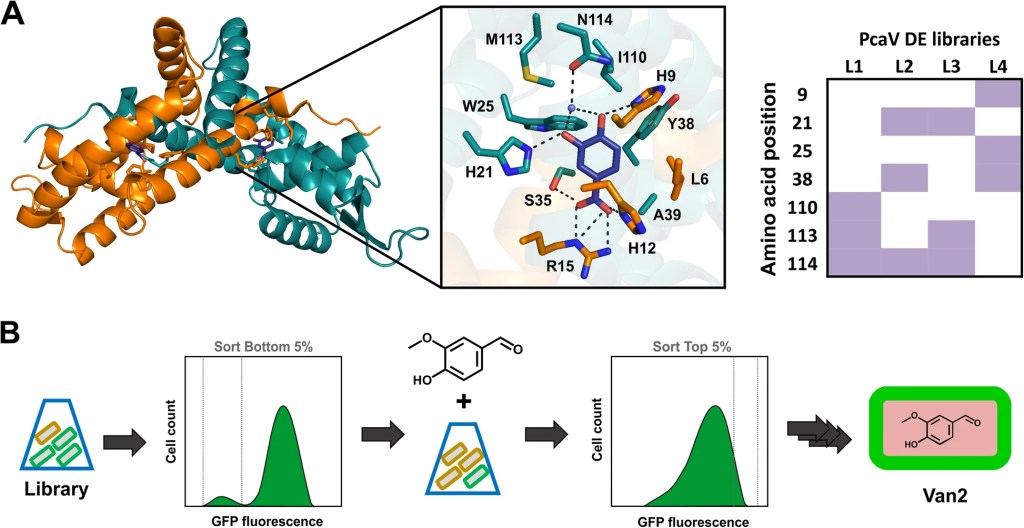
Vanillin biosensor development via directed evolution: The PcaV repressor, a member of the MarR aTF family, was used to develop a biosensor for the detection of hydroxyl-substituted benzoic acids, including protocatechuic acid (PCA). The PCA biosensor was further subjected to directed evolution to alter its ligand specificity towards vanillin and other closely related aromatic aldehydes, to generate the Van2 biosensor. Ligand recognition of Van2 was explored in vitro using a range of biochemical and biophysical analyses, and extensive in vivo genetic-phenotypic analysis was performed to determine the role of each amino acid change upon biosensor performance. (J. Biol Eng 2019)